Project Brief
Contents
5. Project Brief#
A motif is a subgraph with a particular topological structure; for example, there are 13 possible motifs consisting of 3 nodes (see Fig. 5.1). The aim of motif analysis is to construct a motif frequency spectrum (see Fig. 5.2) which describes the relative abundance of each motif within a graph.
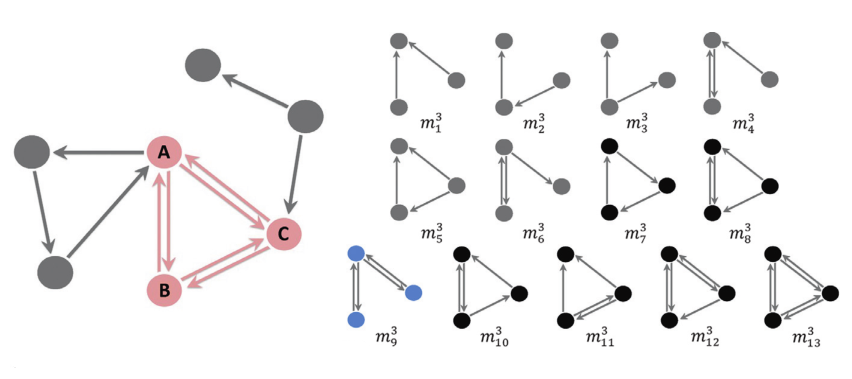
Fig. 5.1 There are 13 distinct three-node motifs in directed networks. Reprinted from [1]#
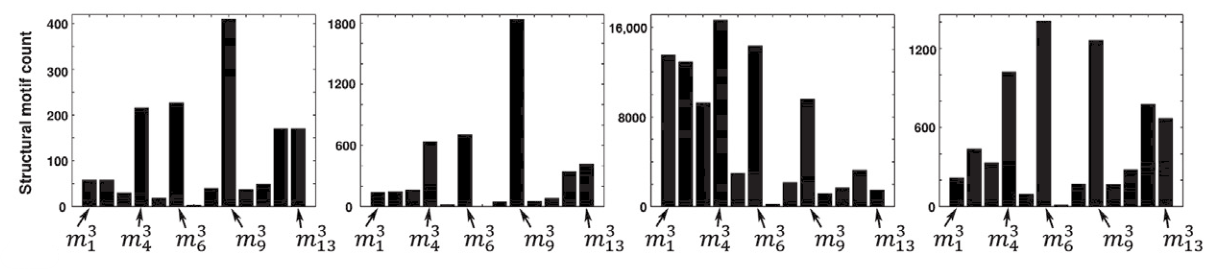
Fig. 5.2 The frequency spectrum of the 13 3-node motifs in four different brain networks. Reprinted from [1]#
5.1. Aims#
In a recent study the connectome of a large part of the Drosophila Melanogaster nervous system was published, consisting of around 25,000 neurons and 20 million synaptic connections (see Fig. 5.3) [3]. The aim of this project is to perform motif analysis by repeatedly sampling from the graph of the drosophila connectome. You will:
Determine the order-3 isomorphic directed graphs
Import the drosophila connectome data and determine how to represent it as a directed graph
Develop code to calculate the motif spectrum of the graph of the drosophila connectome
Compare the resulting graph to a suitable null (reference) model
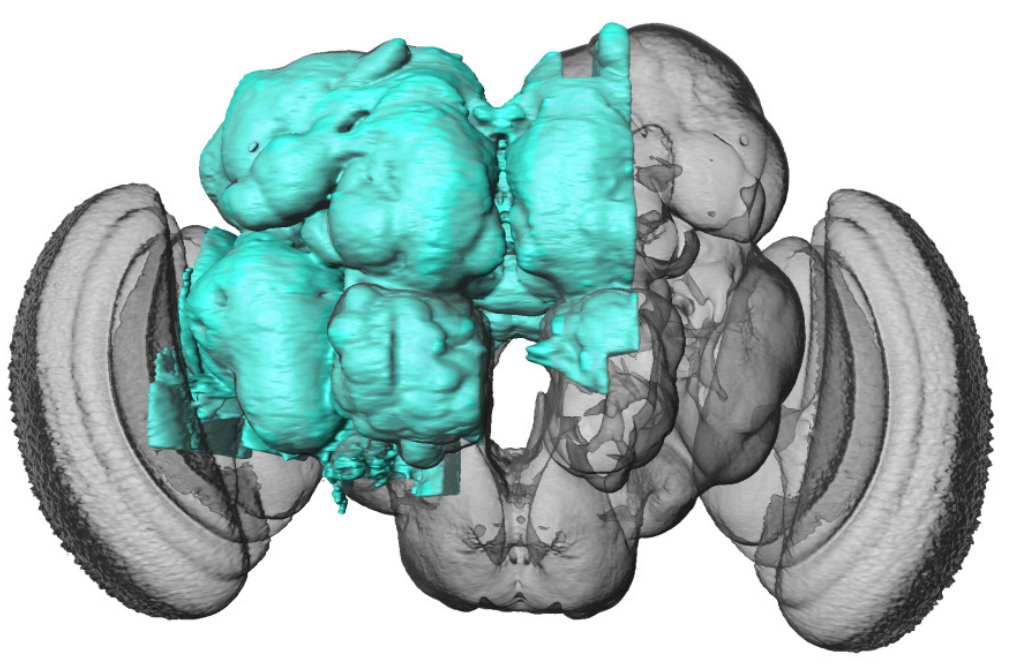
Fig. 5.3 The connectome of the highlighted portion of the drosophila central brain consists of around 25,000 neurons and 20 million synaptic connections. Reprinted from [3]#
5.2. Background#
The suggested text book [1] has some good background and links to further useful information. Chapter 8.1 contains the most relevant material.
You will need to understand just a little more graph theory, in particular the idea of graph isomorphisms, for which notes will be provided.
We will be using a Python library NetworkX to automate some of the graph analysis.
5.3. References#
- 1(1,2,3)
Alex Fornito, Andrew Zalesky, and Edward Bullmore. Fundamentals of brain network analysis. Academic Press, 2016.
- 2
Olaf Sporns and Rolf Kötter. Motifs in brain networks. PLoS biology, 2(11):e369, 2004.
- 3(1,2)
C Shan Xu, Michal Januszewski, Zhiyuan Lu, Shin-ya Takemura, Kenneth J Hayworth, Gary Huang, Kazunori Shinomiya, Jeremy Maitin-Shepard, David Ackerman, Stuart Berg, and others. A connectome of the adult drosophila central brain. BioRxiv, 2020.