with open("Ps_kr_G2B.fasta") as f:
dna_seq = ""
for line in f:
if line[0] != ">":
dna_seq += line.strip()
rna_seq = dna_seq.replace("T", "U")
codon_list = []
n = len(rna_seq)
for i in range(n):
if rna_seq[i:i+3] == "AUG":
j = i + 3
break
for i in range(j, n-3, 3):
codon = rna_seq[i:i+3]
if codon == "UAG" or codon == "UAA" or codon == "UGA":
break
else:
codon_list.append(codon)
genetic_code = ["GCA", "GCC", "GCG", "GCU", "UGC", "UGU", "GAC", "GAU", "GAA", "GAG", "UUC", "UUU", "GGA", "GGC", "GGG", "GGU", "CAC", "CAU", "AUA", "AUC", "AUU", "AAA", "AAG", "UUA", "UUG", "CUA", "CUC", "CUG", "CUU", "AUG", "AAC", "AAU", "CCA", "CCC", "CCG", "CCU", "CAA", "CAG", "AGA", "AGG", "CGA", "CGC", "CGU", "CGG", "AGC", "AGU", "UCA", "UCC", "UCG", "UCU", "ACA", "ACC", "ACG", "ACU", "GUA", "GUC", "GUG", "GUU", "UGG", "UAC", "UAU", "UAG", "UAA", "UGA"]
amino_acids = ["A", "A", "A", "A", "C", "C", "D", "D", "E", "E", "F", "F", "G", "G", "G", "G", "H", "H", "I", "I", "I", "K", "K", "L", "L", "L", "L", "L", "L", "M", "N", "N", "P", "P", "P", "P", "Q", "Q", "R", "R", "R", "R", "R", "R", "S", "S", "S", "S", "S", "S", "T", "T", "T", "T", "V", "V", "V", "V", "W", "Y", "Y", "!", "!", "!"]
protein = ""
for codon in codon_list:
i = genetic_code.index(codon)
aa = amino_acids[i]
protein = protein + aa
print("Protein:", protein)
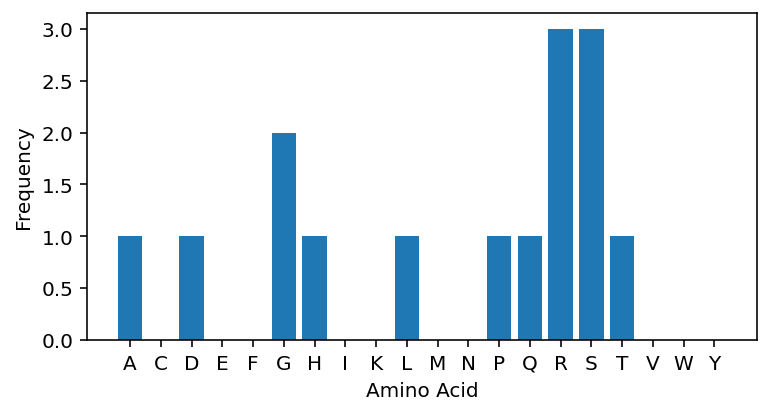
aa = ["A", "C", "D", "E", "F", "G", "H", "I", "K", "L", "M", "N", "P", "Q", "R", "S", "T", "V", "W", "Y"]
aa_counts = [0] * len(aa)
for c in protein:
i = aa.index(c)
aa_counts[i] += 1
import matplotlib.pyplot as plt
plt.figure(figsize=(6,3))
plt.bar(aa, aa_counts)
plt.xlabel("Amino Acid")
plt.ylabel("Frequency");