Chapter 9 exercise
Chapter 9 exercise#
The background setup was given. We just need to change the function f.
The \(x\) and \(y\) ranges are the same.
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
# This is the RHS of the equation
f = lambda x,y: -5*np.sin(3*np.pi*x)*np.cos(2*np.pi*y)
# set up the grid
n=30;
x = np.linspace(0,1,n)
y = np.linspace(0,1,n);
# Useful to store f(x,y) in an array and use F(i,j) instead of f(x(i),y(k))
X,Y = np.meshgrid(x,y)
F=f(X,Y);
# determine grid size
h =x[1]-x[0]
# the solution grid is padded with fictitious nodes to remove at the end
U = np.zeros((n+2,n+2))
# we need to pad F as well, so that the two grids are not mismatched
F=np.pad(F,1, mode='constant')
Then, when we perform the update steps we ensure that the boundary conditions are enforced on the fictitious nodes:
nsweep = 300 #number of sweeps
r=1 #relaxation parameter (can be changed)
n,m = np.shape(U)
for k in range(nsweep-1):
#enforce boundary conditions on fictitious nodes
U[0,:]=U[2,:]; U[-1,:]=U[-3,:]
U[:,0]=U[:,3]; U[:,-1]=U[:,-3]
for i in range(1,n-1):
for j in range(1,m-1):
Unew = (U[i-1,j]+U[i+1,j]+U[i,j-1]+U[i,j+1])/4-h**2/4*F[i,j]
U[i,j] = U[i,j]+r*(Unew-U[i,j])
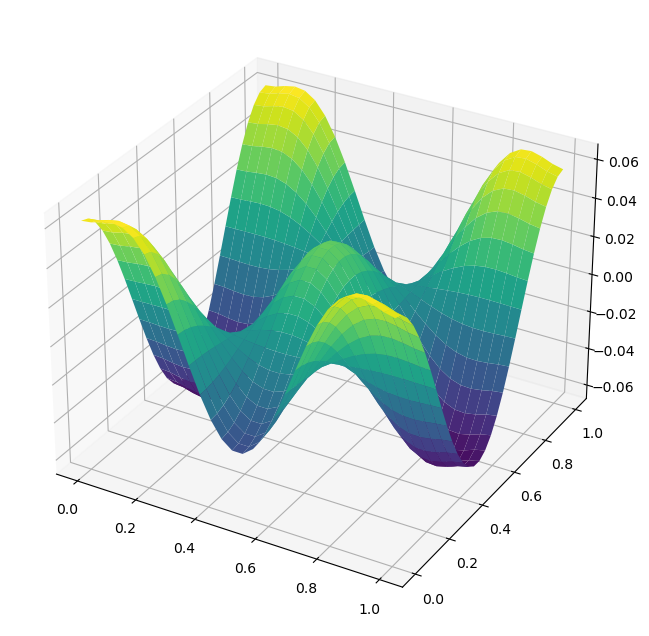
A plot of U[1:-1,1:-1] is shown below. Note that if \(\phi_{sol}\) is a solution of this problem, which satisfies the boundary conditions, then \(\phi_{sol}+C\) is also a solution. Hence, it is possible for the surface to appear shifted up or down in your answers.
fig = plt.figure(figsize=(16, 8))
ax = plt.axes(projection='3d')
ax.plot_surface(X,Y,U[1:-1,1:-1],cmap='viridis')
plt.show()
To further test your understanding, you may wish to try modifying this code so that Neumann conditions on the left and right boundaries are replaced by Dirichlet conditions:
The exact solution to the problem with these conditions is
which will allow you to check your answer.
